 Biography
Biography
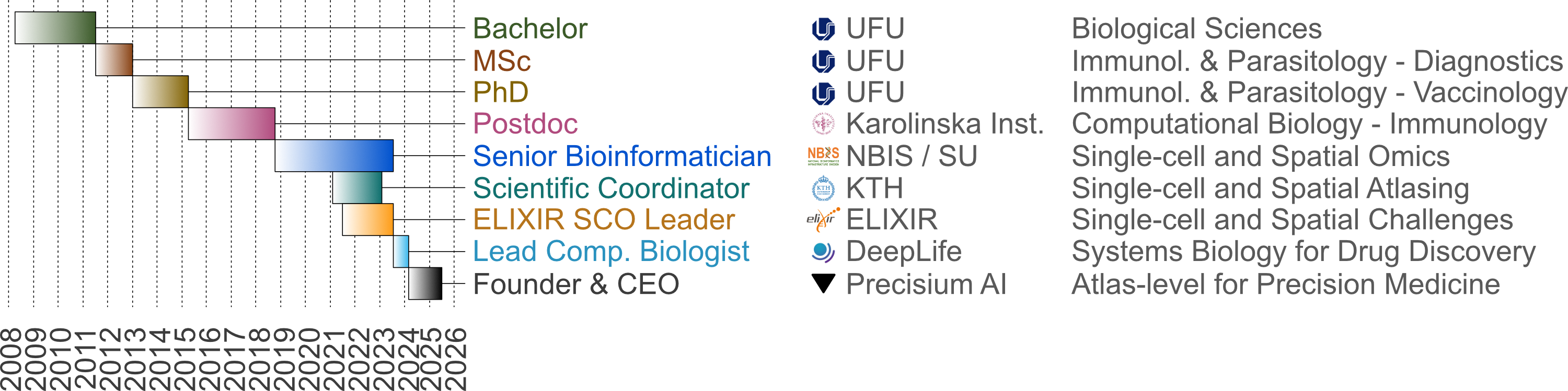
Graphical overview of my career at various stages.
Biography
I enrolled my university studies in 2008 and due to my interest in cell biology in the first university year, I joined the Laboratory of Immunopathology at the Federal University of Uberlandia for a funded scientific internship. This was the laboratory where I completed the Bachelors thesis, Master and finished the PhD by the age of 24. My work focused mainly on Toxoplasma gondii infection and immune responses which lead to the publication of 6 articles (see CV). Since this time, I became interested in data analysis and programming, which led me to apply for a postdoc position in bioinformatics at Karolinska Institutet (KI) in 2015, Sweden.

For the next 3.5 years at the Department of Medicine, I had the opportunity to work on analysis of several high throughput techniques such as bulk and single cell RNA sequencing, proximity extension assay, microarray, 14-color flow cytometry, 16S metagenomics technologies and many others. My work focused on identifying transcriptional signatures in Ulcerative Colitis (UC) patients, which were found to be linked to responsiveness to therapy, and thus provided a way into personalized medicine for UC. This work led to the development of the Atlas of Inflammation and Repair (AIR). I also was temporarily employed by the UKE in Germany to work on single cell data analysis of a rare regulatory cell type, which was found to regulate inflammation in disease patients. Furthermore, due to this diverse analysis portfolio, many international collaborations were established (i.e., US, Germany, France, Belgium, Chile) resulting in highly cited articles. In total, 10 articles were published and 3 others are still under ongoing review.


I then became permanently employed at NBIS in September 2018, and was quickly enrolled in several collaborative projects involving single- or multi-omics modalities coming from single-cell, bulk RNA-seq, in situ sequencing (ISS), among others. Specifically, my work routine focused on ensuring computationally reproducible analysis workflows, developing effective visualization tools for complex data and generating reports to be shared with collaborators, while working on a multitude of projects in parallel. Some consequences of this work were that 1) I helped identify patients with pulmonary sarcoidosis which will respond well to therapy and 2) we discovered a new population state of innate lymphoid cells. In total, 4 articles were published and 4 others are still under ongoing review. Also, teaching and communicating advanced topics such as single-cell data analysis are part of my tasks at NBIS. I have been an active lecturer and course leader in 9 different courses in Sweden and at the European level through ELIXIR, including Finland (CSC) and Switzerland (SIB).

In February 2021, by the age of 29, I was then appointed Scientific Coordinator for the Human Developmental Cell Atlas (HDCA), where I orchestrated and facilitated the interaction of 6 multi-omics research groups. Here, I guide PhD students and Postdocs across different groups to work in synergy and accelerate data analysis and interpretation. Data, code and manuscript curation, as well as giving HDCA visibility and outreach are part of my tasks in this role. Two manuscripts are under ongoing review.
